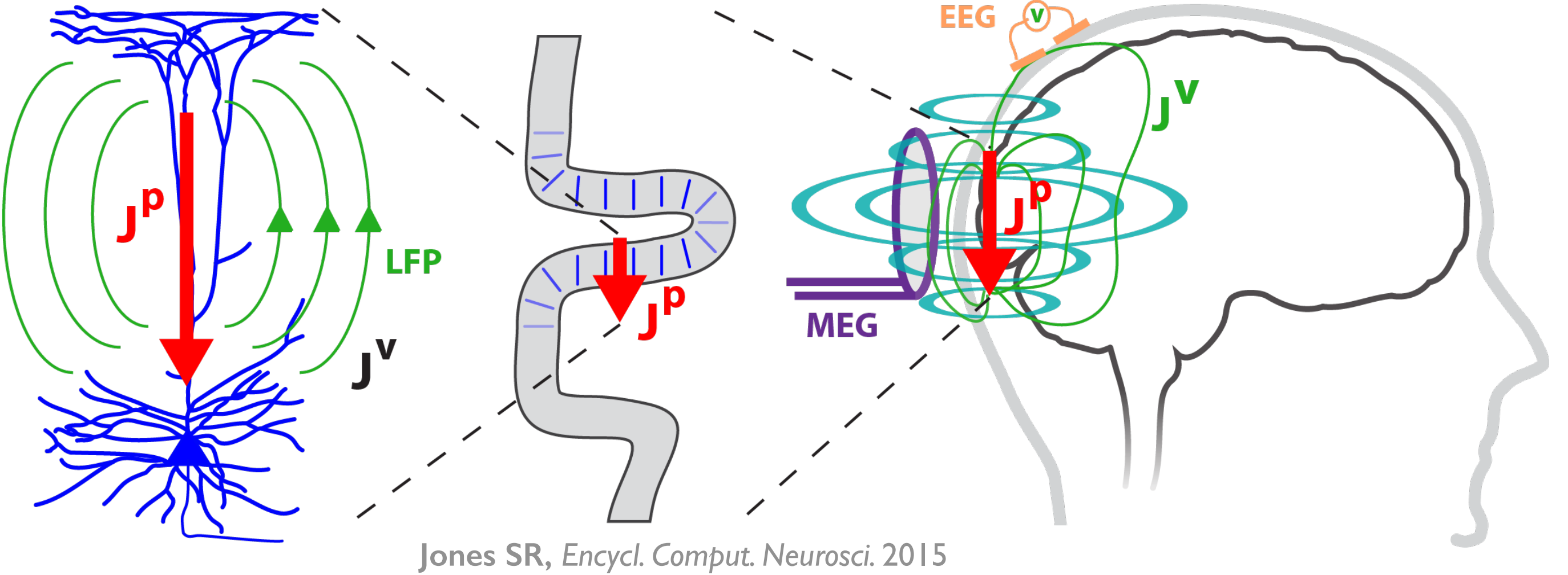
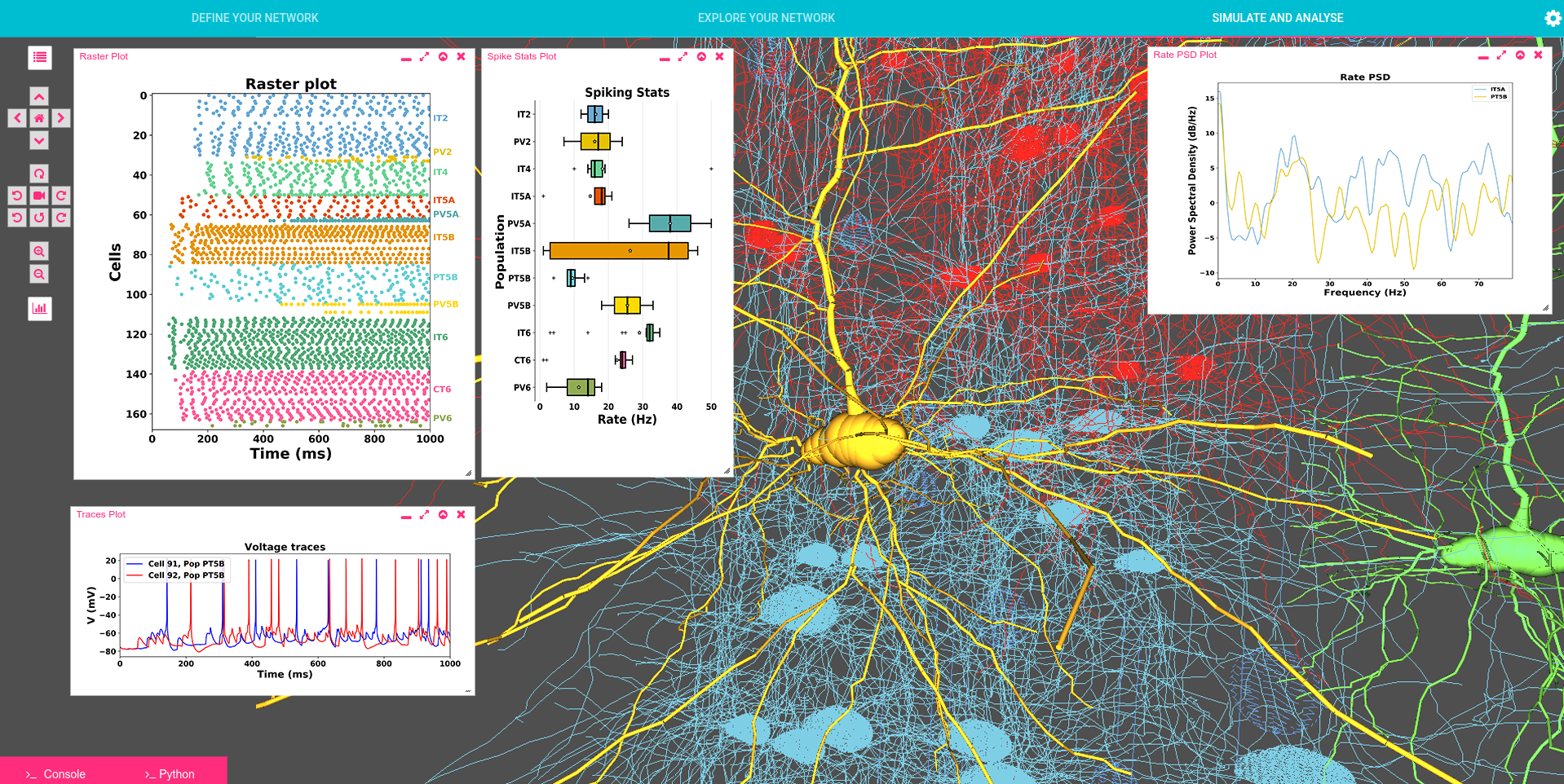
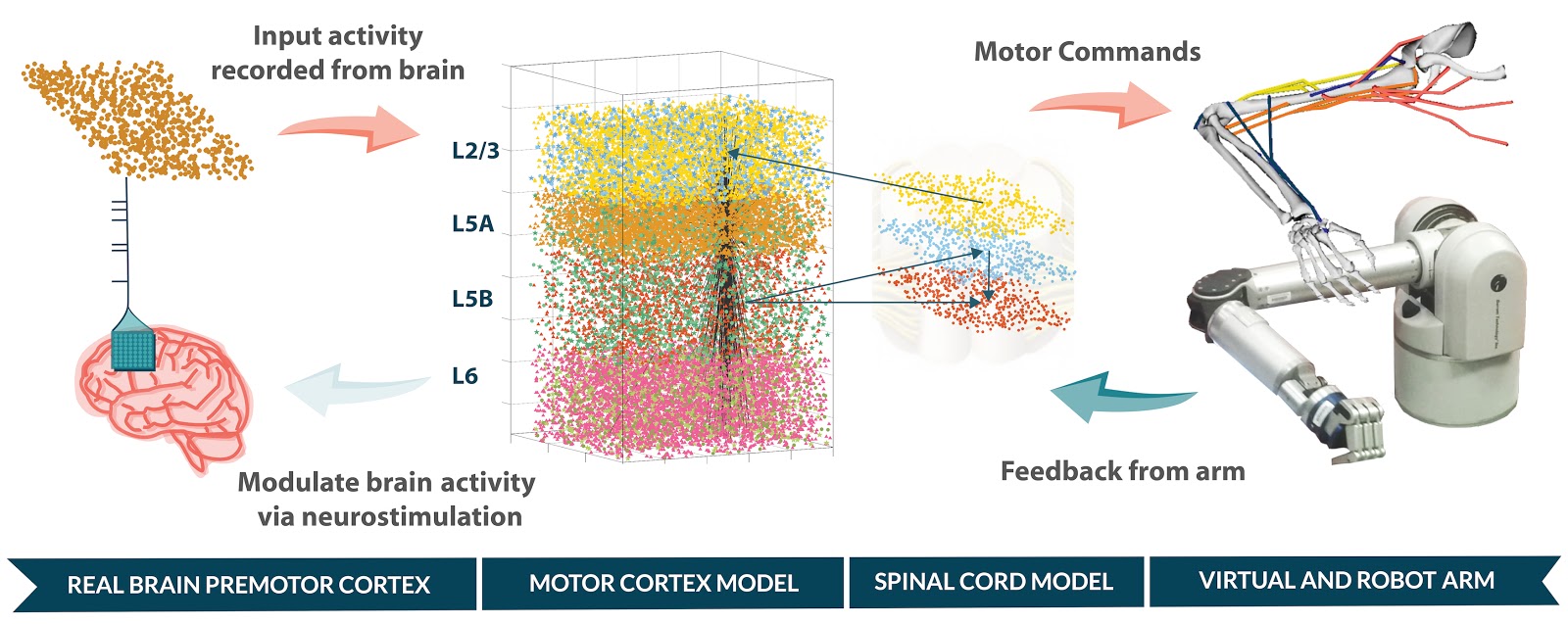
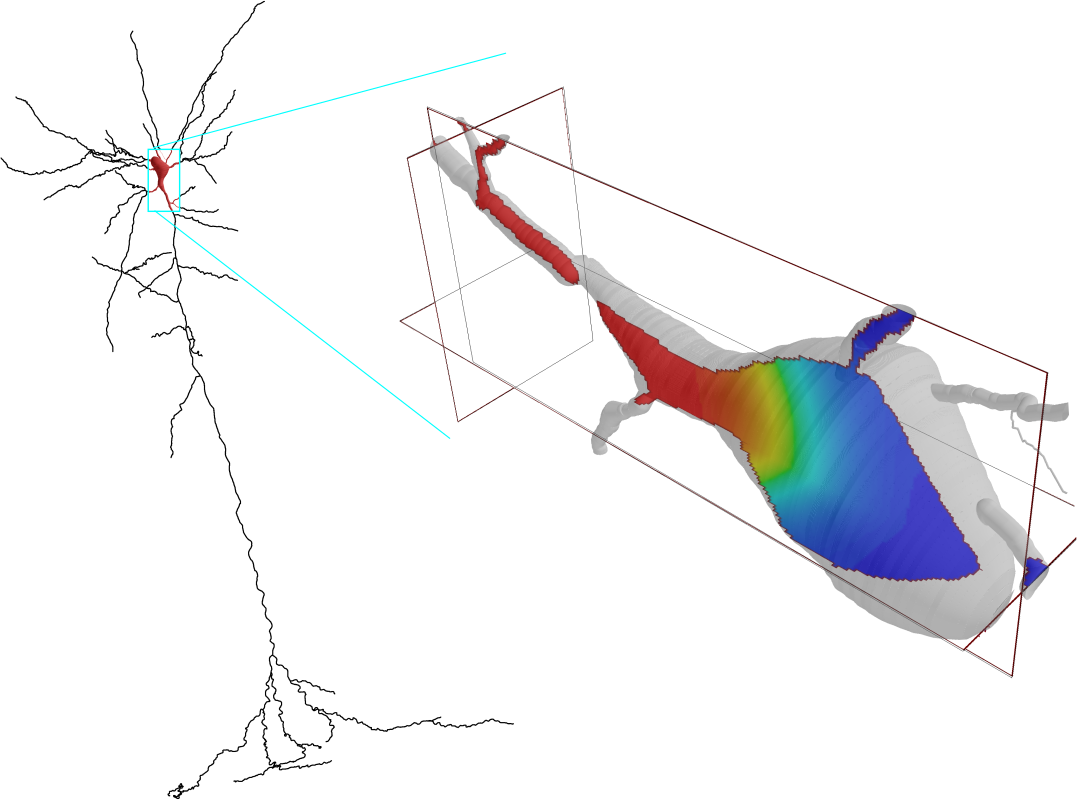
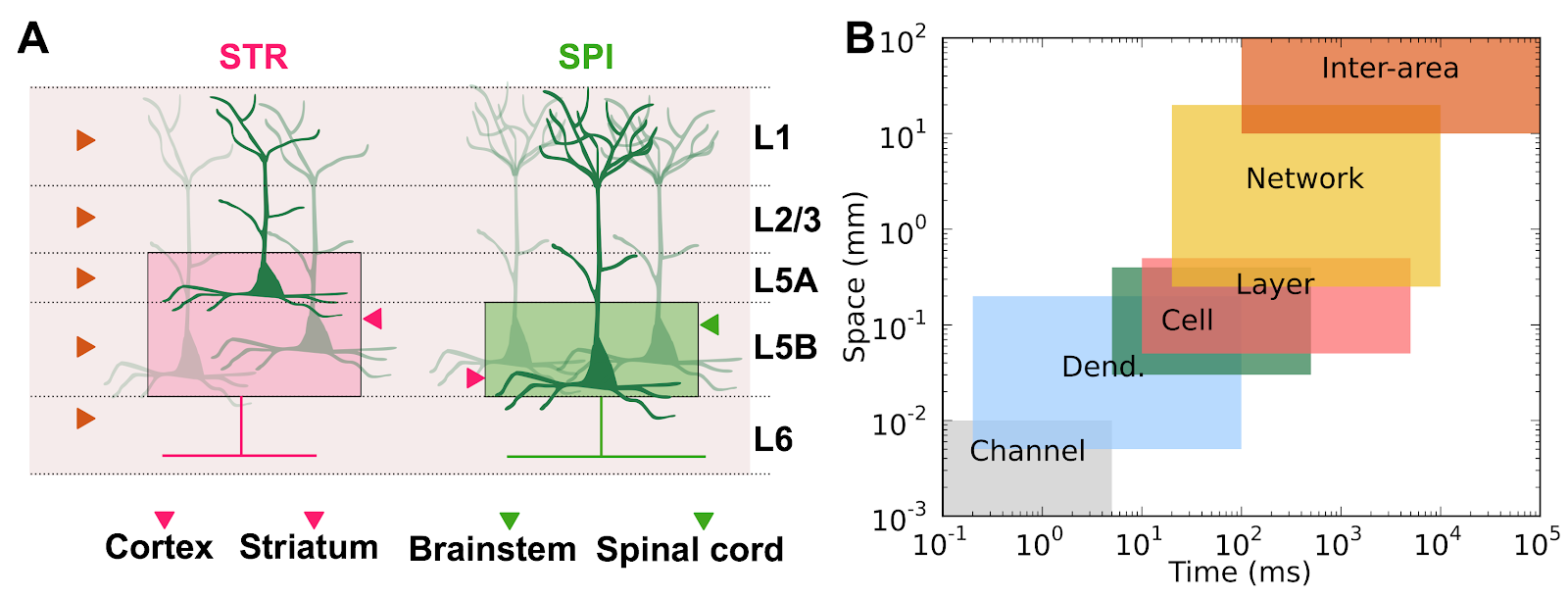
The goal of Human Neocortical Neurosolver (HNN) project is to create a new software tool that gives researchers the ability to develop hypotheses on the location, time-course and circuit-level neural mechanisms underlying human Magneto/Electro-encephalography (MEG/EEG) and ECoG signals. The purpose of the subaward contract with SUNY Downstate is to make use of a unique neuronal network modeling language specification and programming interface (NetPyNE) to substantially increase the utility of the HNN software.